Transcriptomic diversity of Drosophila melanogaster central nervous system
On this website, you can find links to two companion publications in which we explored the transcriptional logic of neuronal identity, and the differences between females and males in that logic, across the central brain at unprecedented detail. We also include links to web apps to plot our single-cell RNA-seq datasets for our central brain neuronal atlas, as well as many subclustered atlases, including doublesex and fruitless clusterings. We hope these apps will allow you to take full advantage of this data by exploring and leveraging it for your own work.
In the first companion paper, we explore the transcriptional diversity across the central brain neurons at unprecedented resolution.
A High-Resolution Atlas of the Brain Predicts Lineage and Birth Order Underly Neuronal Identity.
Cell Genomics. 2025
Allen, A.M., Neville, M.C., Nojima, T., Alejevski, F., Agarwal, D., Sims, D., and Goodwin, S.F.
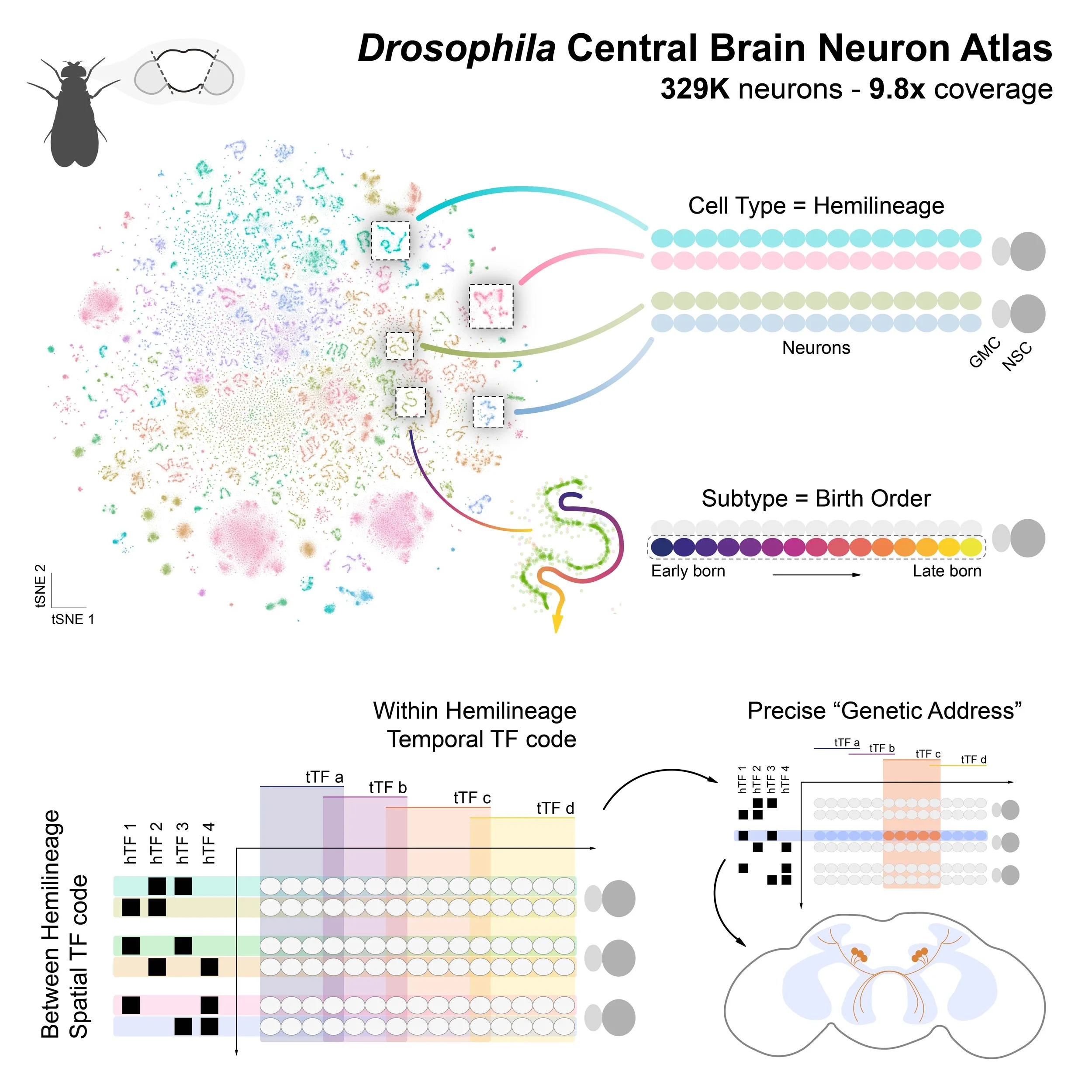
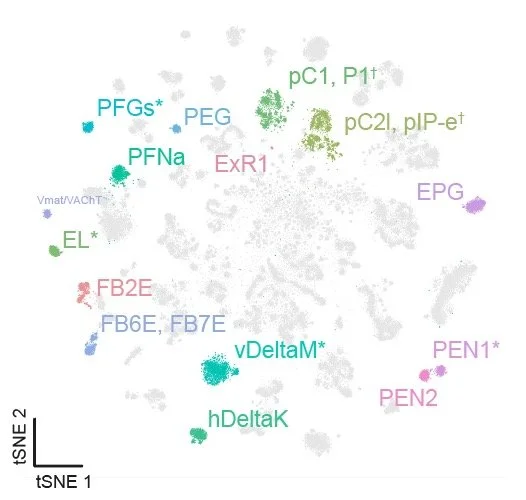
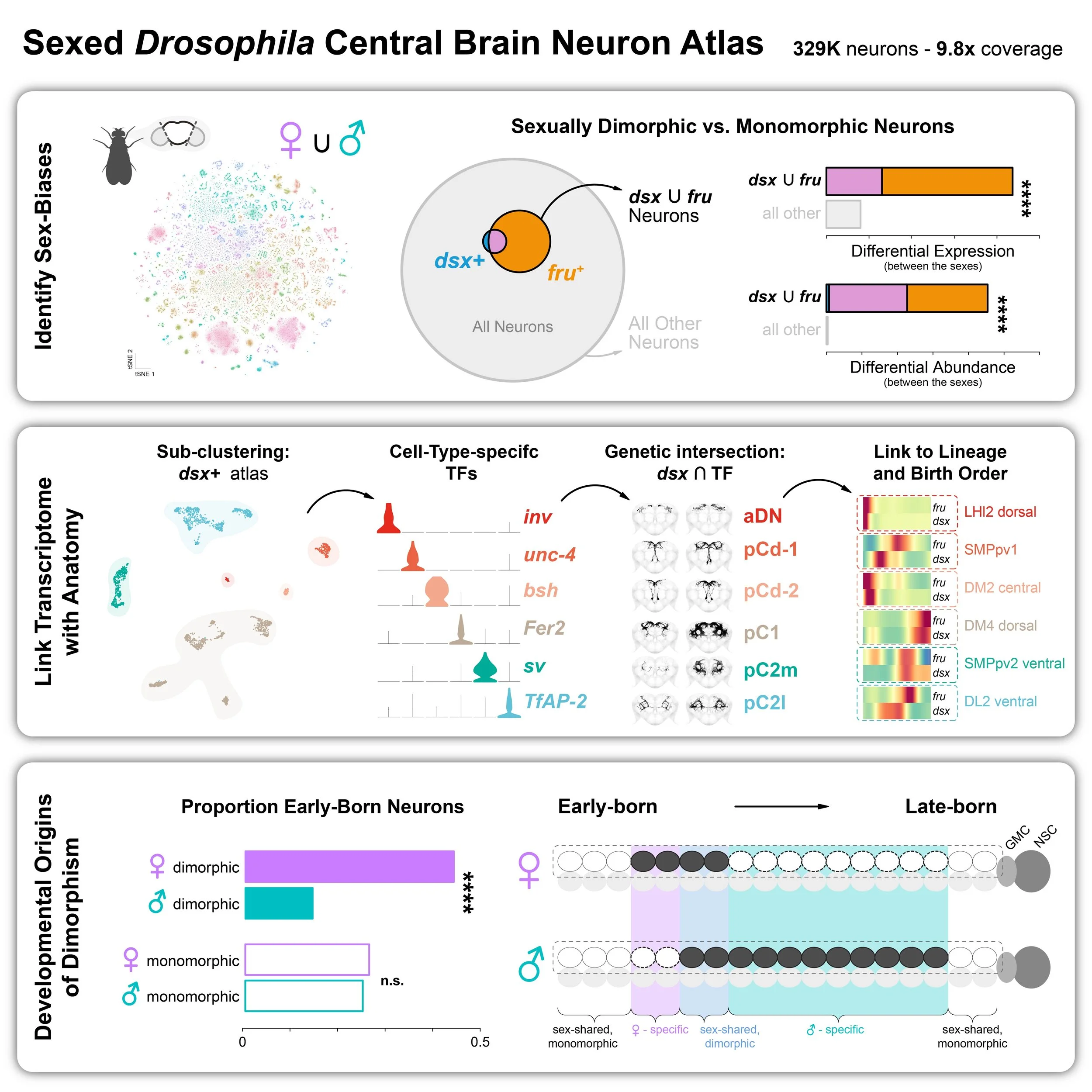
Using an integrated single-cell transcriptomic atlas of the adult Drosophila central brain, Allen et al. uncover over 4,000 transcriptionally distinct neuronal subtypes. The authors show that adult neuronal identity is shaped by a combination of spatial and temporal developmental programs, with distinct transcription factor families acting along each axis. These findings provide a framework for linking gene expression to neural lineage, anatomy, and function.
Highlights
• Single-cell atlas reveals 4,167 neuronal subtypes in adult Drosophila brain.
• Lineage and birth order shape adult neuron transcriptomic identities.
• Distinct TF families act along spatial and temporal axes of neuron specification.
• Intersectional genetic tools bridge gene expression to anatomy and circuit function.
Interactive Web Apps for plotting of the data:
Central Brain Neuronal Atlas:
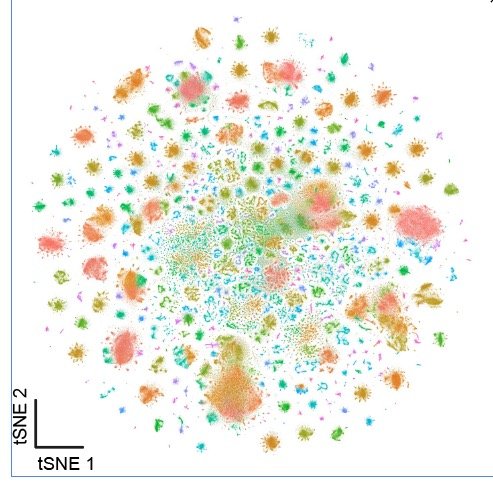
This web app contains plotting features for the central brain neuron atlas. This atlas contains ~330,000 cells and represents a 9.8-fold cellular depth of coverage.
To generate the central brain neuronal atlas, we initially processed a meta-head atlas, then a meta-neuron atlas, and successively filtered out non-neuronal and non-central brain neurons. Those atlases are available below:
Meta Head Atlas:
This web app contains plotting features for the meta-head atlas. This atlas contains more than 1 million cells from various datasets, ranging from whole-head to dissected central brain preparations.
Meta Neuron Atlas:
This web app contains plotting features for the meta-neuron atlas. This atlas contains more than 700,000 neurons from various datasets, ranging from whole-head to dissected central brain preparations.
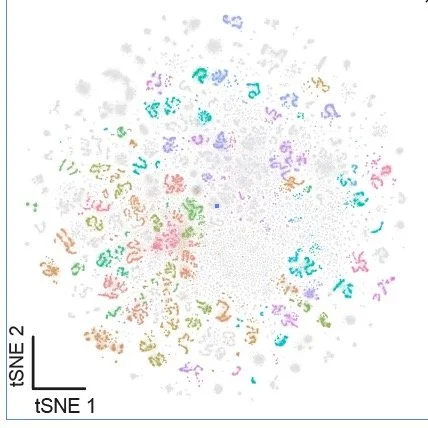
Central Brain Neuronal Atlas: (Kenyon Cells Removed)
This web app contains plotting features for the central brain neuron atlas with the Kenyon cells removed. The large number of Kenyon cells can obscure the visual clarity when looking at other cell types, so we have removed them in this web app.
Kenyon Cell Atlas:
This web app contains plotting features for the Kenyon cell atlas. These neurons were removed from the previous object and can be visualized here.
Type II Neuronal Atlas:
This web app contains plotting features for the type II neuron atlas. Type II neurons give rise to many central complex neurons, as well as to many sexually dimorphic neuronal populations.
Endocrine Neuron Atlas:
This web app contains plotting features for the endocrine neuron atlas, as identified by the expression of dimm.
Monoaminergic Neuron Atlas:
This web app contains plotting features for the monoaminergic neuron atlas, as identified by the expression of Vmat.
Monoaminergic neurons: (PAMs removed)
This web app contains plotting features for the monoaminergic neuron atlas, with the PAMs removed.
PAM neurons:
This web app contains plotting features for the monoaminergic PAM neuron atlas.
In the second companion paper, we explore the differences in these transcriptional landscapes between females and males.
Differential Neuronal Survival Defines a Novel Axis of Sexual Dimorphism in the Drosophila brain.
Cell Genomics. 2025
Allen, A.M., Neville, M.C., Nojima, T., Alejevski, F., and Goodwin, S.F.
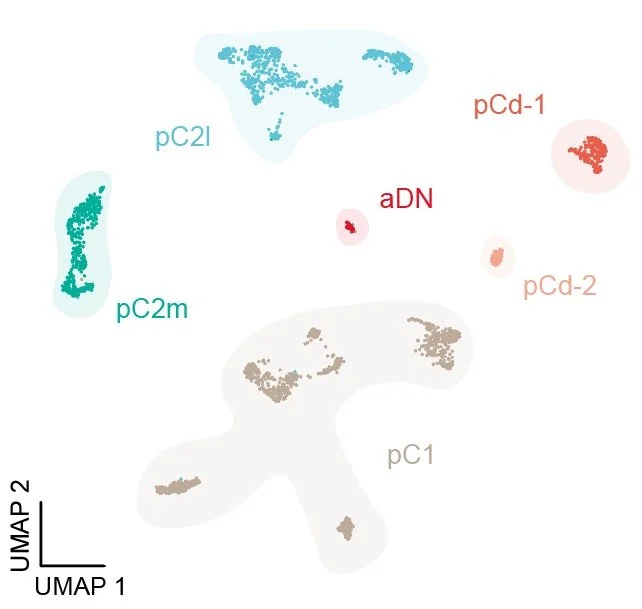
Allen et al. leverage a high-resolution single-cell transcriptomic atlas of the adult Drosophila central brain to map sexual dimorphism. They show that dimorphic neuronal types correlate with the expression of the sex-determination transcription factors Doublesex and Fruitless. The authors uncover extensive transcriptional diversity within these dimorphic cell types and link their molecular profiles to anatomical identities. They further identify birth order as a previously unrecognized axis of dimorphism, driven by sex-biased neuronal survival.
Highlights
• Systematic analysis of sex-specific transcriptomic diversity in the adult brain
• doublesex and fruitless explain the majority of sex differences in the adult brain
• Linking doublesex and fruitless transcriptomic diversity to neuron anatomy
• Birth order represents a novel axis of sexual differentiation in the central brain
Interactive Web Apps for plotting of the data:
Central Brain Neuronal Atlas:
This web app contains plotting features for the central brain neuron atlas. This atlas contains ~330,000 cells and represents a 9.8-fold cellular depth of coverage.
doublesex Neuronal Atlas:
This web app contains plotting features for the doublesex neuron atlas.
fruitless Neuronal Atlas:
This web app contains plotting features for the fruitless neuron atlas. Kenyon cells and monoaminergic neurons were removed from this subclustering and are treated separately.
doublesex and fruitless Union Neuronal Atlas:
This web app contains plotting features for the doublesex and fruitless union neuronal atlas. Neurons in this atlas express doublesex, fruitless, or both (Kenyon cells and monoaminergic neurons were excluded).
doublesex and fruitless Shared Lineage Neuronal Atlas:
This web app contains plotting features for the doublesex and fruitless shared lineage neuronal atlas. The above union atlas was subclustered to only contain neurons from hemilineages with shared doublesex and fruitless expression. Neurons in this atlas express doublesex, fruitless, or both.